Research Interests & Experience
I am broadly interested in computer vision, data compression, and novel applications of deep learning. My current work leverages Gemini models to empower wearable AI with ego-centric vision, enabling agents to better understand and interact with the physical world. Specifically, I am leading the development of long-term memory and navigational capabilities on smart glasses. Previously, I was the Comfort Quality Lead at Google AR, where I designed and led evaluation frameworks to optimize user comfort and video see-through quality for head-mounted displays, including the first Android XR headset - Samsung Galaxy XR.
Publications


Human Hands as Probes for Interactive Object Understanding
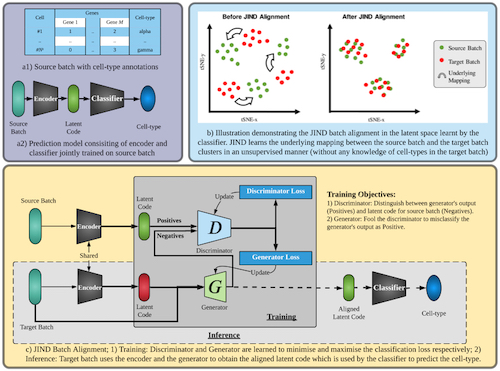
JIND: Joint Integration and Discrimination for Automated Single-Cell Annotation